Current Research Themes 研究テーマ
Below is a brief description on the current research activities in our laboratory with related references and publications for further readings and more technical details. We are open to discuss new ideas and research plans of related topics. In that case, please consult with the director.
以下は、研究室での現在の研究テーマの簡単な説明であり、関連する論文とともに、さらなる読み物や技術的な詳細があります。 関連するトピックに関する新しいアイデアや研究計画がある場合は、教授に相談してください。
以下は、研究室での現在の研究テーマの簡単な説明であり、関連する論文とともに、さらなる読み物や技術的な詳細があります。 関連するトピックに関する新しいアイデアや研究計画がある場合は、教授に相談してください。
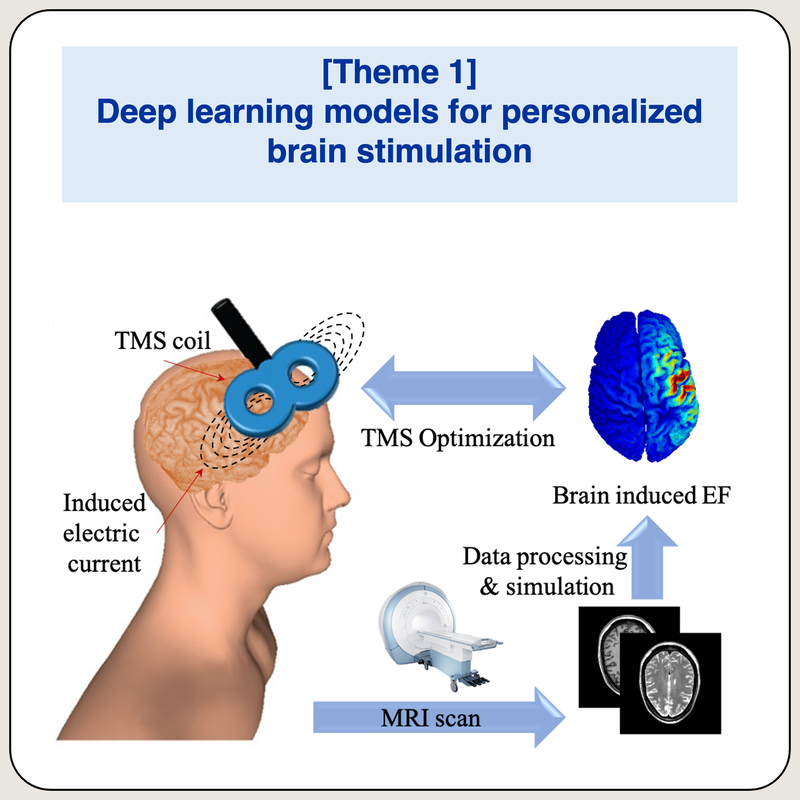
Deep learning models for personalized brain stimulation
|
We study how deep learning can be employed to solve different problems associated with brain stimulation. Given the fact that brain (and also non-brain) tissues are essential in electromagnetic studies, we were the first to use deep learning architecture (named, ForkNet) to generate a full head segmentation from magnetic resonance imaging (MRI) scans into 13 different head tissues. By using ForkNet, a personalized full head model can be generated from MRI data in few seconds. We also invented a novel approach for automatic estimation of dielectric properties directly from anatomical images based on the estimation of water contents. The main idea is based on realistic assumption of non-uniform dielectric properties of living tissues. This research direction aims at the development of more feasible deep learning techniques that can be used in clinical applications.
Publications & References: [1] https://doi.org/10.1109/ACCESS.2023.3268133 (IEEE Access, IF22=3.9, 2023) [2] https://doi.org/10.1109/TEMC.2022.3212860 (IEEE TEMC, IF22=2.1, 2022) [3] https://doi.org/10.1088/1361-6560/ac7b64 (Phys. Med. Biol., IF22=3.5, 2022) Review [4] https://doi.org/10.3389/fnins.2021.695668 (Front. Neurosci., IF22=4.3, 2021) [5] https://doi.org/10.1088/1361-6560/abe223 (Phys. Med. Biol., IF22=3.5, 2021) [6] https://doi.org/10.1016/j.neunet.2020.02.006 (Neural Nets., IF22=7.8, 2020) [7] https://doi.org/10.1088/1361-6560/ab7308 (Phys. Med. Biol., IF22=3.5, 2020) [8] https://doi.org/10.1109/TMI.2020.2969682 (IEEE Trans. Med. Imag., IF22=10.6, 2020) [9] https://doi.org/10.1016/j.neuroimage.2019.116132 (NeuroImage, IF22=5.7, 2019) [10] https://doi.org/10.1109/MPULS.2019.2923888 (IEEE Pulse, IF22=0.6, 2019) Tutorial |
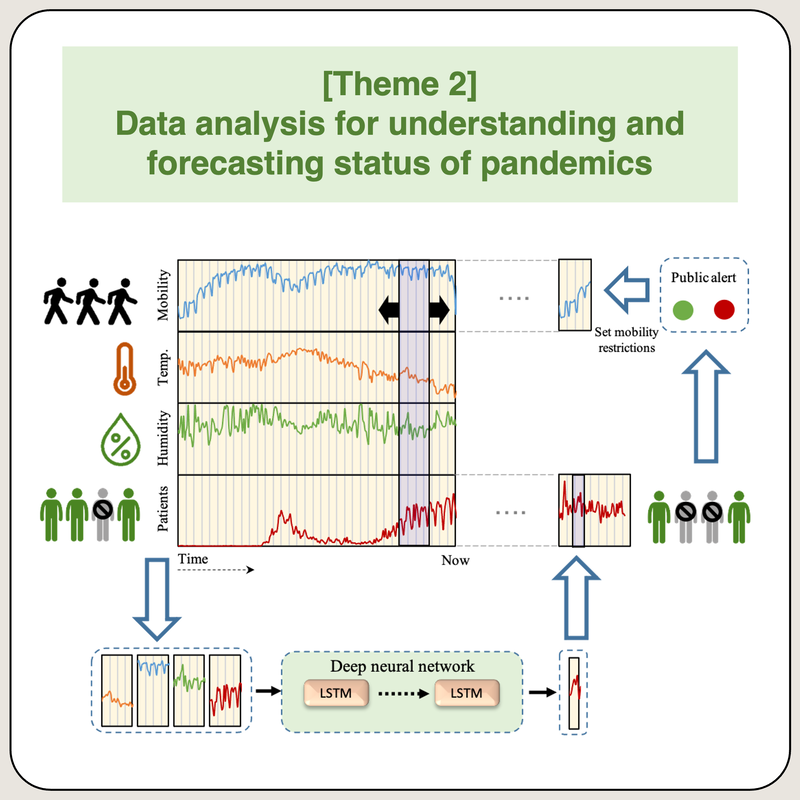
Data analysis for understanding and forecasting status of pandemicS
|
The outbreak of the coronavirus disease (COVID-19) had been reported worldwide. Commonly, the morbidity and mortality rates of COVID-19 differ by one or more orders of magnitude in each country. However, proper comparison is not straightforward because of various co-factors, such as human behavior and regional policies; a similar problem occurs with the determination of the morbidity and mortality rates. We investigate using different deep learning models to forecast the future of the pandemic with different mixture of available data. This process includes data analysis, feature extraction and model design. Results of our past research were included in the national COVID-19 AI and Simulation Project coordinated by Japanese Cabinet Secretariat (https: //www.covid19-ai.jp). Also our results appear in newspapers and TV interviews.
Publications & References: [1] https://doi.org/10.3390/vaccines11030633 (Vaccines, IF22=7.8, 2023) [2] https://doi.org/10.1007/s11524-022-00697-5 (J. Urban Health, IF22=6.6, 2022) [3] https://doi.org/10.3390/vaccines10111820 (Vaccines, IF22=7.8, 2022) [4] https://doi.org/10.1016/j.compbiomed.2022.105986 (Comp. Biol. Med., IF22=7.7, 2022) [5] https://doi.org/10.1016/j.compbiomed.2022.105548 (Comp. Biol. Med., IF22=7.7, 2022) [6] https://doi.org/10.3390/vaccines10030430 (Vaccines, IF22=7.8, 2022) [7] https://doi.org/10.1016/j.jbi.2021.103743 (J. Biomed. Info., IF22=4.5, 2021) [8] https://doi.org/10.1016/j.onehlt.2020.100203 (One Health, IF22=5.0, 2021) [9] https://doi.org/10.3390/ijerph18157799 (Int. J. Env. Res. Pub. Health, IF21=4.614, 2021) [10] https://doi.org/10.3390/ijerph18115736 (Int. J. Env. Res. Pub. Health, IF21=4.614, 2021) [11] https://doi.org/10.3390/ijerph17155477 (Int. J. Env. Res. Pub. Health, IF21=4.614, 2020) [12] https://doi.org/10.3390/ijerph17155354 (Int. J. Env. Res. Pub. Health, IF21=4.614, 2020) |
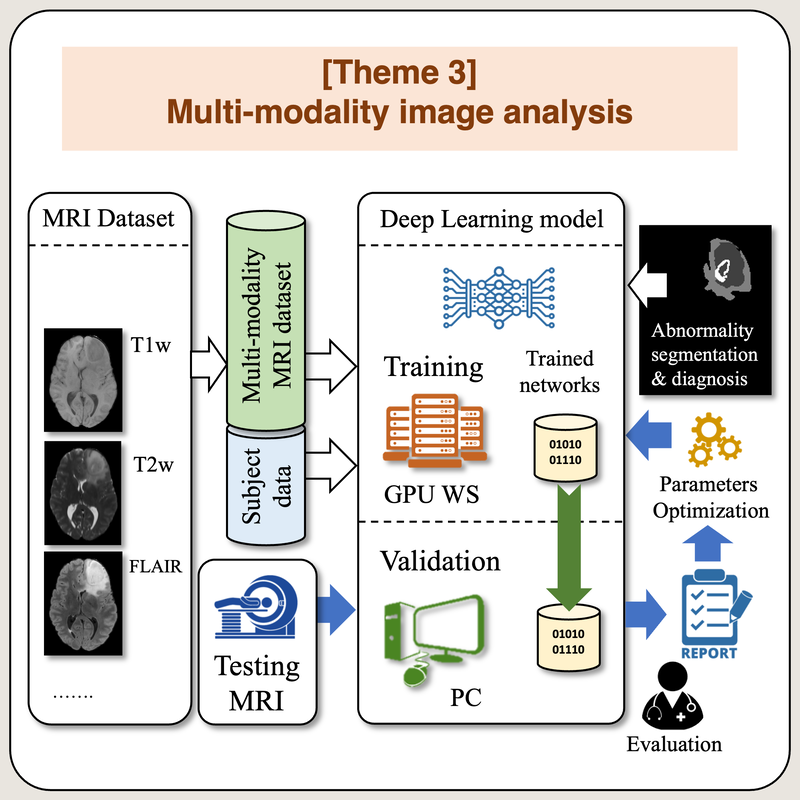
Multi-modality image analysis
|
Understanding medical image through visual observation of multi-modal images may improve understanding of image data and would enable better diagnosis in several clinical applications. However, looking at different images is a time consuming task that is difficult to be well-processed with limited expert manpower. The aim of this research theme is to design, develop and implement deep learning models for multi-modal image analysis especially for cancer diseases.
Publications and References: [1] https://doi.org/10.1186/s43055-023-01014-z (EJRNM, IF22=1.0, 2023) [2] https://doi.org/10.1371/journal.pone.0276523 (Plos One, IF22=3.7, 2022) [3] https://doi.org/10.1371/journal.pone.0262349 (Plos One, IF22=3.7, 2022) |
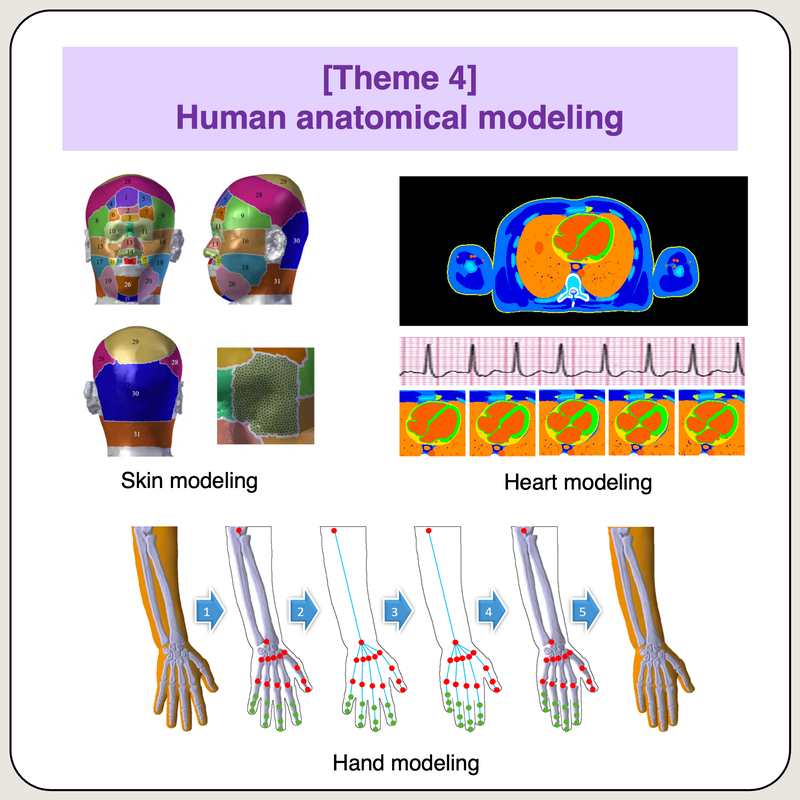
Human anatomical modeling
|
Human anatomical models are digital objects that is commonly used in simulation studies for different applications including imaging, human safety, drug design, therapy planning, etc. Several digital objects (models/phantoms) are available in static form that can not represent physiological effects such as human breathing and normal movements. In this research, we develop several methods to generate reliable dynamic human models that can efficiently represent real objects for better simulation studies and more accurate experimental results.
Publications and References: [1] https://doi.org/10.48550/arXiv.2309.06677 (arXiv preprint, 2023) [2] https://doi.org/10.1109/TEMC.2022.3212860 (IEEE TEMC, IF22=2.1, 2022) [3] https://doi.org/10.3390/s21134275 (Sensors, IF22=3.9, 2021) [4] https://doi.org/10.1109/ACCESS.2020.3035815 (IEEE Access, IF22=3.9, 2020) [5] https://doi.org/10.1109/ACCESS.2020.3017773 (IEEE Access, IF22=3.9, 2020) [6] https://doi.org/10.1109/TEMC.2020.2985404 (IEEE TEMC, IF22=2.1, 2020) [7] https://doi.org/10.1016/j.compbiomed.2020.104009 (Comp. Biol. Med., IF22=7.7, 2020) [8] https://doi.org/10.1109/ACCESS.2019.2904743 (IEEE Access, IF22=3.9, 2019) |